import pandas as pdPandas Basic Analysis 01
01 Basic Data Analysis with Python Pandas
Content Outline
- Introduction
- Load data from a text file
- Display summary information about the data
- Data analysis using the groupby() operation
- Display data plots
- Filtering data rows using the loc() method
Introduction
Pandas is a Python package used extensively in data manipulation and analysis. The purpose of this Colab notebook is to give an overview of Pandas. Subsequent notebooks in this tutorial series elaborate on various aspects of Pandas.
import matplotlib
import matplotlib.pyplot as pltLoad Data From a Text File
For this demonstration, we’ll work with global demographic data sourced from the Gapminder Foundation. Before proceeding, we load the tab-delimeted data file “gapminder.tsv” to Google Colab. The data is available from this Github repo.
df = pd.read_csv("sample_data/gapminder.tsv", sep='\t')Display Summary Information About the Data
A cursory look at the data shows that it has six columns and 1704 rows. In Pandas, a set of row and columnar data is known as a data frame. Every data frame comes with an info() method that breaks down the tabulated data. The keys() method of a data frame returns a list of indices, or column names. To enumerate the column data themselves, a data frame’s values attribute returns the information as a two-dimensional array.
df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1704 entries, 0 to 1703
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 country 1704 non-null object
1 continent 1704 non-null object
2 year 1704 non-null int64
3 lifeExp 1704 non-null float64
4 pop 1704 non-null int64
5 gdpPercap 1704 non-null float64
dtypes: float64(2), int64(2), object(2)
memory usage: 80.0+ KBdf.keys()Index(['country', 'continent', 'year', 'lifeExp', 'pop', 'gdpPercap'], dtype='object')df.valuesarray([['Afghanistan', 'Asia', 1952, 28.801, 8425333, 779.4453145],
['Afghanistan', 'Asia', 1957, 30.332, 9240934, 820.8530296],
['Afghanistan', 'Asia', 1962, 31.997, 10267083, 853.10071],
...,
['Zimbabwe', 'Africa', 1997, 46.809, 11404948, 792.4499603],
['Zimbabwe', 'Africa', 2002, 39.989, 11926563, 672.0386227],
['Zimbabwe', 'Africa', 2007, 43.487, 12311143, 469.7092981]],
dtype=object)That was the structure of the data frame. Now let’s analayze the data itself. The Pandas describe() method reports the standard statistics for the data frame.
df.describe()| year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|
| count | 1704.00000 | 1704.000000 | 1.704000e+03 | 1704.000000 |
| mean | 1979.50000 | 59.474439 | 2.960121e+07 | 7215.327081 |
| std | 17.26533 | 12.917107 | 1.061579e+08 | 9857.454543 |
| min | 1952.00000 | 23.599000 | 6.001100e+04 | 241.165876 |
| 25% | 1965.75000 | 48.198000 | 2.793664e+06 | 1202.060309 |
| 50% | 1979.50000 | 60.712500 | 7.023596e+06 | 3531.846988 |
| 75% | 1993.25000 | 70.845500 | 1.958522e+07 | 9325.462346 |
| max | 2007.00000 | 82.603000 | 1.318683e+09 | 113523.132900 |
Data Analysis Using the Groupby() Operation
Let’s go deeper. Say we want to find the average life expectancy by year. Here’s how we instruct Pandas to retrieve that information.
df.groupby('year')['lifeExp'].mean()year
1952 49.057620
1957 51.507401
1962 53.609249
1967 55.678290
1972 57.647386
1977 59.570157
1982 61.533197
1987 63.212613
1992 64.160338
1997 65.014676
2002 65.694923
2007 67.007423
Name: lifeExp, dtype: float64One way of understanding what just happened is that Pandas separates and analzyes the data as buckets, each one representing a year. Judging by the result above, not all years are present in our data as we only see twelve non-contiguous years between 1952 and 2007.
Display Data Plots
Next we plot the graph for this data. Note that if we were to do so in the Python or iPython interactive console, we’d have to first import a Python display library, Qt for example, for displaying the graph as a GUI window; then we register it to matplotlib just before we run the show() method: * import PyQt5 * matplotlib.use(‘Qt5Agg’)
df.groupby('year')['lifeExp'].mean().plot()
plt.show()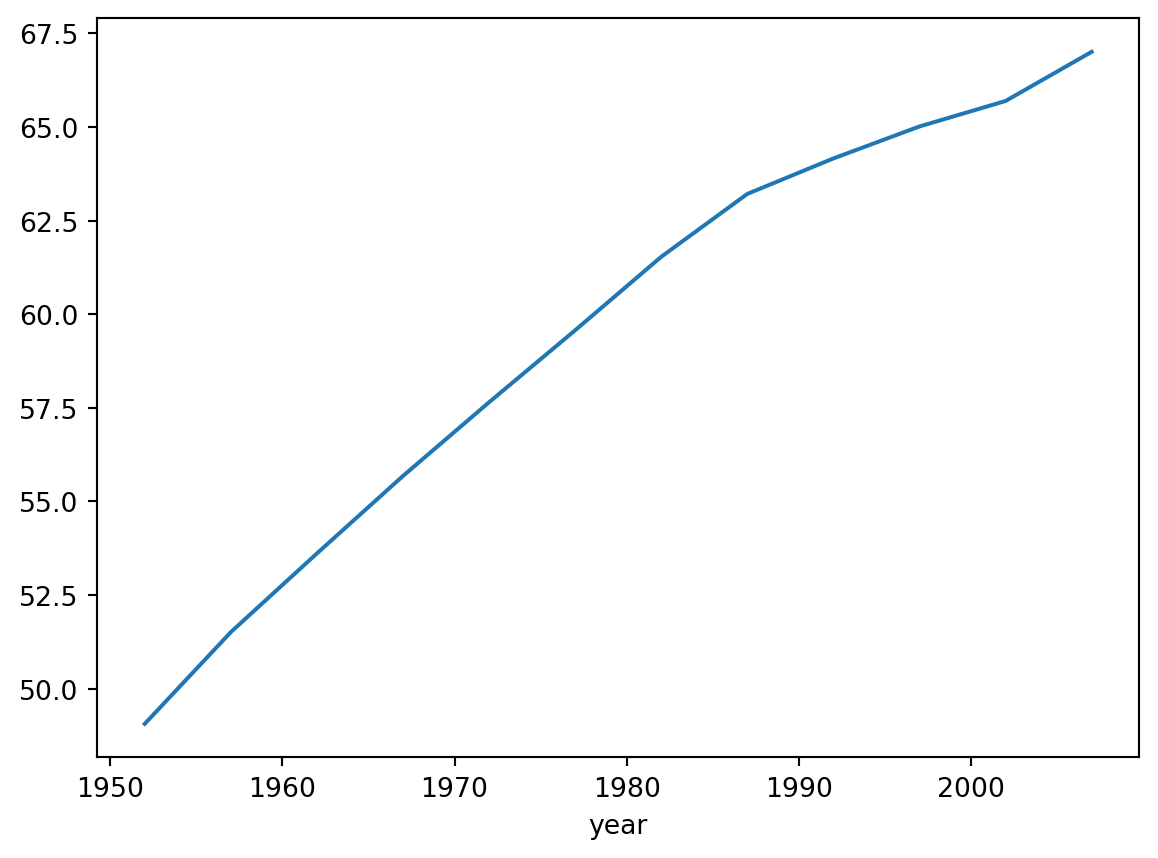
Now let’s go a bit further. Say we want to find the life expentancy data grouped by year and by country.
df.groupby(['year', 'country'])[['pop', 'lifeExp']].mean()| pop | lifeExp | ||
|---|---|---|---|
| year | country | ||
| 1952 | Afghanistan | 8425333.0 | 28.801 |
| Albania | 1282697.0 | 55.230 | |
| Algeria | 9279525.0 | 43.077 | |
| Angola | 4232095.0 | 30.015 | |
| Argentina | 17876956.0 | 62.485 | |
| ... | ... | ... | ... |
| 2007 | Vietnam | 85262356.0 | 74.249 |
| West Bank and Gaza | 4018332.0 | 73.422 | |
| Yemen, Rep. | 22211743.0 | 62.698 | |
| Zambia | 11746035.0 | 42.384 | |
| Zimbabwe | 12311143.0 | 43.487 |
1704 rows × 2 columns
Filtering Data Rows Using the loc() Method
This is a lot of information to take in and Pandas only presents a snapshot of the 1704 rows. To make it easier for ourselves, we can ask Pandas to limit the information to a list of years, say 1972, 1982 qne 1987. We use Panda’s loc() method to filter rows based on their label.
df.groupby(['year', 'country'])[['pop', 'lifeExp']].mean().loc[[1972, 1982, 1987]]| pop | lifeExp | ||
|---|---|---|---|
| year | country | ||
| 1972 | Afghanistan | 13079460.0 | 36.088 |
| Albania | 2263554.0 | 67.690 | |
| Algeria | 14760787.0 | 54.518 | |
| Angola | 5894858.0 | 37.928 | |
| Argentina | 24779799.0 | 67.065 | |
| ... | ... | ... | ... |
| 1987 | Vietnam | 62826491.0 | 62.820 |
| West Bank and Gaza | 1691210.0 | 67.046 | |
| Yemen, Rep. | 11219340.0 | 52.922 | |
| Zambia | 7272406.0 | 50.821 | |
| Zimbabwe | 9216418.0 | 62.351 |
426 rows × 2 columns
It’s worth discussing a few fine points about Panda’s concept of a data row. Before we grouped the data by year, a row was a unit of information in the gapminder.tsv data file. That is, the original Pandas data frame was made up of all 1704 rows. However, now that we applied the groupby() method, Pandas organized the data such that a row bundles information by year. In this new, derived data frame, a row is now indexed by the year: 1972, 1982 and so on. Therefore when we apply the loc() method, we’re filtering these subset of grouped-by rows, not the original rows in the data file.
Applying the loc() method a number of times, we can futher refine the rows of information we extract for a particular year.
df.groupby(['year', 'country'])[['pop', 'lifeExp']].mean().loc[1972].loc[['Albania', 'Brazil', 'Vietnam']]| pop | lifeExp | |
|---|---|---|
| country | ||
| Albania | 2263554.0 | 67.690 |
| Brazil | 100840058.0 | 59.504 |
| Vietnam | 44655014.0 | 50.254 |
